Engineered Sequences
Gene technologies—such as transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeats (CRISPRs), or zinc finger nucleases (ZFNs)—have grown exponentially in recent years.
While these technologies have created new opportunities in areas like food supply and medicine, their ability to modify genes poses a significant bioterrorism threat to the nation. Law enforcement and defense agencies must find a way to identify genes that have been engineered, as well as ways to counteract potentially harmful effects. Using our proprietary BioVelocity tool, Noblis has succeeded in identifying genetically modified, potentially harmful organisms. We are also researching engineered sequence technology to help identify methods for counteracting the effects of gene mutations.
Screening Engineered Sequences with BioVelocity
The latest generation of genome editing tools has simplified the process of modifying eukaryotic genomes, raising the concern that bad actors could create new classes of modified pathogens for misuse. Noblis has invested significant internal research funds to develop bioinformatics capabilities that can identify and characterize potential mutations. BioVelocity, a proprietary Noblis bioinformatics tool, can be used to:
- Screen large volumes of genomic DNA sequence information significantly faster than available competing commercial off-the-shelf bioinformatics capabilities
- Determine whether genetic anomalies are the result of a CRISPR-driven modification event
- Determine whether genetic anomalies are single nucleotide polymorphisms, an indel (insertion or deletion), or part of an engineered sequence
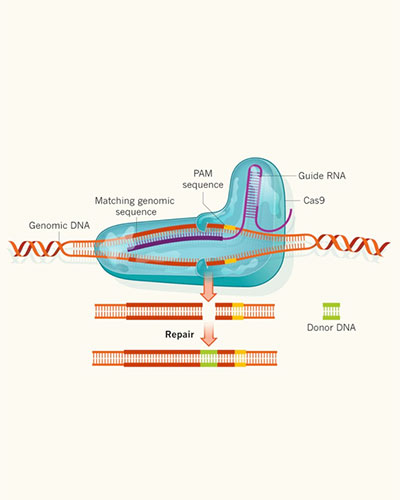
Structure-Based Protein Engineering and Design
Noblis uses three-dimensional modeling of peptides as part of our bioinformatics analysis to predict phenotype/functional impacts of engineering and mutation. Our tools leverage crystal and solved structures of large molecules to identify changes in hydrophobicity and solvent accessible surface after a sequence change. This key data point allows our scientists to predict if a change would yield a functional variant. From this new surface, we use the Adaptive Poisson-Boltzmann Solver to calculate electrostatics and new bonding locations and angles resulting from the engineering or mutation.
This research capability enhances our understanding of:
- The nature and specificity of receptor-ligand interactions for developing enhanced drug therapeutics
- Protein structure for designing more efficient, thermo-tolerant, or altered-substrate enzymes
- Immunogen antigenicity for designing more efficacious vaccine candidates
Assessing the Ecological Impact of Gene Drives
Gene drive is a means to ensure 100% inheritance of a new gene into a population. Using gene drive technology for biological control to eliminate undesirable mosquito populations or create pest-resistant crops is an exciting and potentially powerful application of synthetic biology to address pressing consumer needs and concerns. However, little information is available concerning the ecological impact of releasing engineered populations into a naïve environment. Noblis has an ethical interest in ensuring that we protect the environment and minimize potential risk to our ecosystem resulting from such activities. Noblis will work with customers to:
- Mathematically model potential fate and impact of modified populations on an environment to support regulatory assessments
- Discuss and develop reverse-gene-drive options as safeguard measures to counter potential negative consequences of an accidental release of genetically modified strains
- Help develop and execute field test plans to generate statistically valid data to verify predictive models in support of environmental release studies

Engineered Sequences – Reducing the Problem Set
Identifying novel and unknown engineered sequences in a pool of many unknown gene sequences is a challenge for those tasked with biological defense. Not every organization will be able to bring high-performance computing to bear to tackle this challenge. Noblis is researching novel techniques and algorithms to allow researchers to focus on those reads or k-mers that are more likely to contain engineered sequences.
We are leveraging probabilistic data structures, such as Bloom Filters and Count-Min Sketches, to determine which sequences are more likely to be engineered.
The goals of this research are to:
- Reduce the computational resources necessary to detect and identify engineered sequences - processing and memory ideally on a consumer laptop
- Reduce time from raw whole genomic sequencing to analysis
Additional Resources
The In-Q-Tel laboratory for emerging technologies in the life sciences: diagnostics, vaccines, biothreats, analytics.
bnext.org
IARPA Research program: FELIX: Finding Engineering-Linked Indicators
www.iarpa.gov/index.php/research-programs/felix